Document Type : Original Article
Authors
1 Department of Pharmaceutical Chemistry, Faculty of Pharmacy, Near East University, Lefkosa TRNC, Mersin10, Turkey
2 Department of Endodontics, Faculty of Dentistry, Near East University, Lefkosa TRNC, Mersin10, Turkey
3 Department of Pharmacology, Faculty of Pharmacy, Near East University, Lefkosa TRNC, Mersin10, Turkey
4 Department of Pharmacology, Faculty of Dentistry, Near East University, Lefkosa TRNC, Mersin10, Turkey
Abstract
The NLRP3 (NOD-like receptor family containing a pyrin domain 3) inflammasome pathway has a crucial role in the dental immune system and is associated with the activation of the dental immune response. Therefore, it is a specific target for drug molecules to be selected in the treatment of endodontic diseases. Various NLRP3 inflammatory and caspase-1 inhibitors that exhibit effective inhibition against inflammatory conditions have been identified in previous studies. In this study, the human NLRP3 model was constructed by the loop modeling method using computer-aided programs. Binding affinities, inhibition constants (Ki), and ligand-protein interactions of the selected ligands were calculated and investigated by molecular docking simulation against the inflammasome NLRP3 and caspase-1. Binding modes and calculations were performed according to Lamarckian genetic algorithm. The calculated docking scores for each ligand used in this study were between the range of -5.1 and -11.8 kcal/mol for the inhibitory activity. CY-09 (a NLRP3 inflammasome inhibitor) and VX-765 (a caspase-1 inhibitor) were shown to have the most desirable binding affinities, Ki values, and strong binding interactions in the NLRP3 and human caspase-1 binding pockets, respectively. The combination of CY-09 and VX-765 ligands can be used to prevent inflammation in the treatment of endodontic diseases. These inhibitors could be used in the future treatment of endodontic infections and to improve the viability of root canal drugs and pulp capping materials.
Graphical Abstract
Keywords
Main Subjects
Introduction
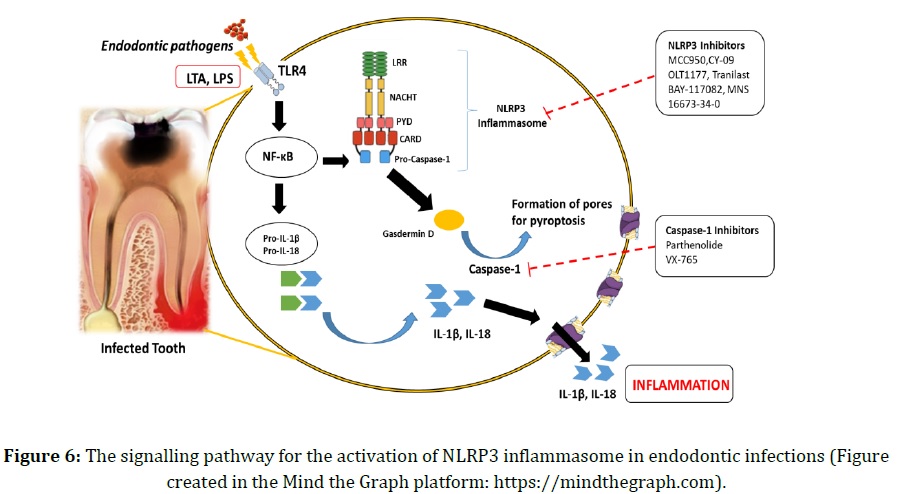
Endodontic disease is defined as the immunoinflammatory response of the pulp or periapical tissues against endodontic pathogens exhibiting various virulence factors associated with host infection [1]. Lipopolysaccharides (endotoxin, LPS) and lipoteichoic acid (LTA) are considered as the most important virulence factors because they have the highest immunogenicity in terms of pulp or periapical tissue damage [2]. Innate immune cells possess surface receptors known as pattern recognition receptors (PRRs) that recognize two subclasses of molecules termed pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs). The NOD-Like receptors (NLRs), one of the cytosolic PRRs, recognize PAMP signals and can form an inflammatory complex called inflammasome [3]. The inflammasome is a protein complex that recognizes stimuli with potential to cause inflammation and, in response, initiates the production of pro-inflammatory cytokines (IL-1β and IL-18) through the activation of caspase-1 [4].
Furthermore, caspase-1 generates pro-Gastermin-D, and then transmembrane Gastermin-D pores are formed to induce cell death [5]. Although there are different types of inflammasome structures, the NLRP3 inflammasome (NOD-like receptor family containing a pyrin domain 3) has been reported to be associated with pulpitis and apical periodontitis [5]. Therefore, NLRP3 is involved in the dental immune system and is activated by many bacterial stimuli [6]. The NLRP3 consists of 3 main parts; the N-terminal pyrine effector domain (PYD), the NACHT domain [NAIP (neuronal apoptosis inhibitor protein), CIITA (an activator of transcription protein), HET-E (a locus protein), and TP1 (telomerase-associated protein)] as well as one Leucine-rich repeat domain (LRR) [7]. The LRR domain, located at the carboxy terminus, recognizes microbial ligands and endogenous hazardous molecules [7]. The NACHT domain is involved in the oligomerization of the NLRP3 inflammasome and consists mainly of two motifs termed Walker A and Walker B [8]. The Walker A site (P-loop) is responsible for the ATPase activity. Since it is an important part of the ATP-binding site, it could be used as a target for in silico studies of new NLRP3 inhibitors [8].
LPS and LTA can stimulate Toll-like receptors (TLR4 type) that activate the nuclear factor kappa light chain enhancer of activated B cells (NFκB) and promote expression of NLRP3, IL-1β, and IL-18 [9]. A recent study has highlighted that the NLRP3 inflammasome plays a crucial role in triggering the dental immune response and may represent a specific target for the treatment of endodontic diseases [10].
Therefore, the aim of this molecular docking study was to evaluate the binding energies and interactions of different inhibitors known to play an important role in inflammation through the direct interaction of NLRP3 binding sites or through the inhibition of caspase-1, and also to select the most effective combination for use in the endodontic treatment or as a complementary combination for the main strategy of endodontic treatment. Our selected inhibitors could be used simultaneously to prevent inflammasome complex formation, caspase-1 activity, and pro-Gasdermin-D cleavage leading to cellular pyroptosis and a range of inflammatory responses aggravating the endodontic diseases.
Materials and Methods
Preparation of data set
A human NLRP3 model and docking calculation based on the crystal structure of rabbit nucleotide-binding oligomerization domain-containing protein 2 (NOD2) in the ADP-bound state (PDB ID: 5IRM) and the alignment insertions closer to the binding site were built by loop modelling using Modeller 9.15 software [11], and then the created model was prepared for molecular docking. On the other hand, the crystal structure of the peptide NLRP3-PYD (PDB ID: 3QF2) and the crystal structure of Caspase-1 (PDB ID: 1RWK) were downloaded from the Protein Data Bank (PDB) of the Research Collaboratory for Structural Bioinformatics (RCSB). A selection of specific ligands for each macromolecule was performed prior to the docking process. The previously reported NLRP3 inflammasome inhibitors, CY-09 (PubChem CID 44561595) [12], 3,4-methylenedioxy-β-nitrostyrene (MNS) (PubChem CID 672296) [13], OLT1177 (Dapansutrile) (PubChem CID 12714644) [14], Oridonin (PubChem CID 5321010) [15], Tranilast (PubChem CID 5282230) [16], and MCC950 (PubChem CID 9910393) [17] were preferred for the molecular docking study of the human NLRP3 model. On the other hand, the specific inhibitors of the NLRP3-PYD domain, BAY 11-7082 (PubChem CID 5353431) [18] and 16673-34-0 (PubChem CID 85542) [19], were chosen as ligands for the NLRP3-PYD peptide. Furthermore, the previously described caspase-1 inhibitors [5], Parthenolide (PubChem CID 7251185) and Belnacasan (VX-765) (PubChem CID 11398092) were used as ligands for the caspase-1 docking process. Before docking, the structures of all ligands were downloaded from the PubChem database and converted into the mol2 file format using the Open Babel 3.1 software.
Structure refinement, molecular docking and interaction analysis
For structure refinement, all water molecules and hetero-atoms were removed from the protein structure, polar hydrogens were added, non-polar hydrogens were merged, and Gasteiger charges were assigned. After refinement, they were converted into PDBQT file format via AutoDockTools4 [20]. Autodock 4.0 software was used for molecular docking studies to predict the ligand binding modes selected in its associated target macromolecule. For the ligands to be energetically favourable and for the correct arrangement of the molecules in space to be determined, the energy should be minimized. For the minimization process, the AutoDock software uses the Lamarckian genetic algorithm [21]. A conformal cluster analysis was performed allowing for a root mean square deviation (RMSD) of 2.0 Å on the docked results. The docking study was performed separately to evaluate the selected antagonists known to inhibit the designed human NLRP3 model, NLRP3-PYD domain, and Caspase-1 protein. The grid box sizes were set to 25 × 25 × 28 A◦ for all docking studies. The most favourable docking poses were represented by the binding affinity (ΔG). Subsequently, the inhibition constant (Ki) and the molecular interaction of the selected poses were evaluated. The structures of proteins, ligands, and docking complexes were visualized using the Discovery Studio 3.0 software. When molecular docking is performed with a rigid receptor, the accurate visualization of ligand and receptor interactions may not always be achieved. To overcome this limitation, multiple molecular docking programs can be used with different algorithms and molecular dynamics simulations.
Results and Discussion
In general, the binding affinities of all ligands were favorable between the range -5.1 and -11.8 kcal/mol. On the other hand, the predicted Ki values of all ligands were in the nanomolar (nM) range, indicating good binding properties, since ligands with smaller inhibition constants bind more tightly to their target macromolecule [22]. Thus, the lower the Ki values, the higher the binding affinity of the ligands.
The interacting residues, types of interactions, binding affinities, and inhibition constant values of the ligands 16673-34-0 and BAY117082 against the peptide NLRP3-PYD are summarized in Table 1.
The docking results show that both ligands bind to the type I site of the NLRP3-PYD peptide, mainly through hydrogen bonding, alkyl and π-alkyl interactions as shown in Figure 1.
In addition, VX-765, a caspase-1 inhibitor, appears to have better binding affinity compared with another caspase-1 inhibitor, Parthenolide, as listed in Table 2. In the docking process of VX-765 and Parthenolide, the common amino acid residues involved in the interaction in the caspase-1 binding pocket were revealed as Arg383, His342, Met345, Val348, and Gly346 with electrostatic interactions as shown in Figure 2.
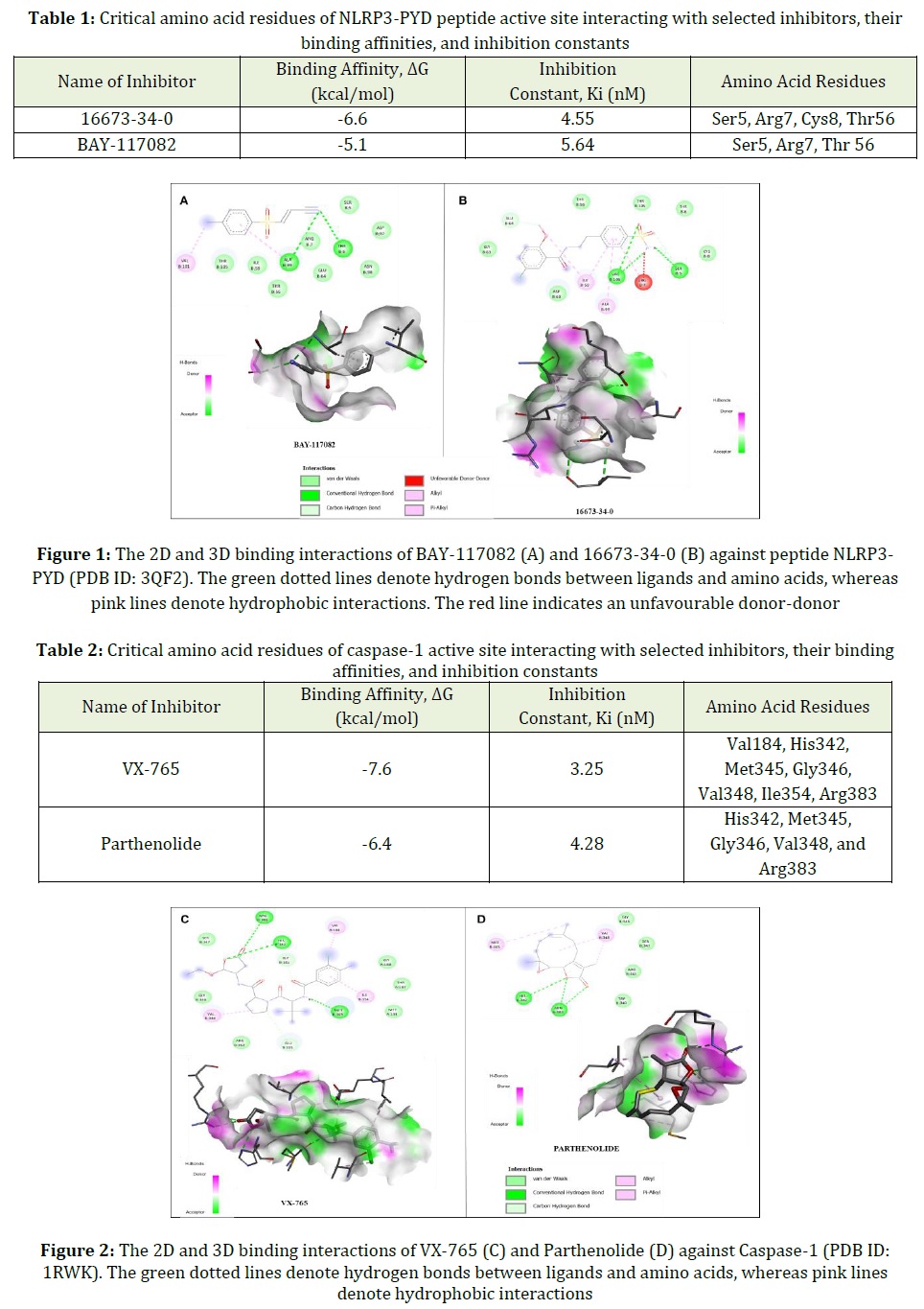
In addition, MCC950, Tranilast, Oridonin, MNS, and OLT1177 were found to interact with human NLRP3 binding sites mainly through electrostatic forces. The 2D representation of molecular interactions for these ligands is shown in Figures 3 and 4.
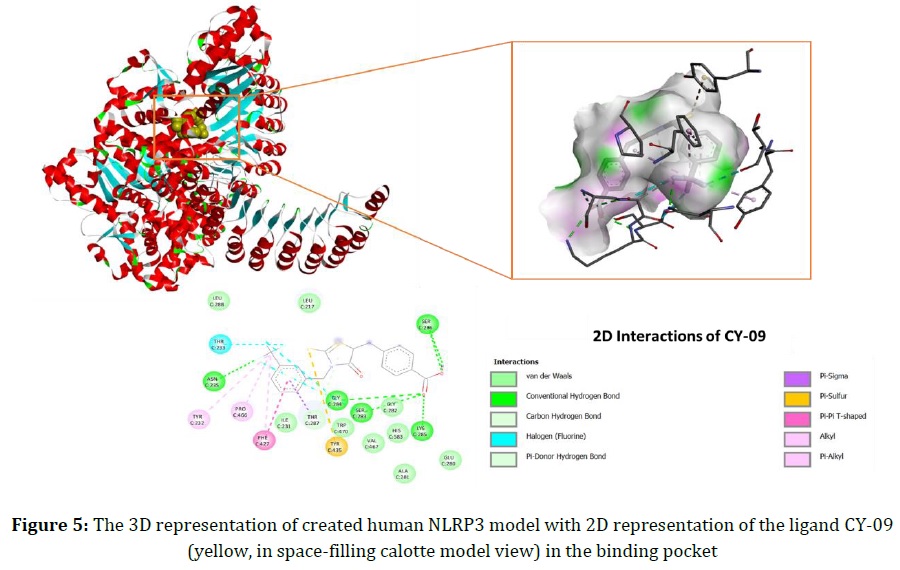
Among other ligands, CY-09 has better binding affinity and inhibition constant as summarized in Table 3. Aside from hydrogen bonding, alkyl, and π-π stacking interactions, CY-09 generates a π-sulfur interaction with Tyr435 and a halogen (fluorine) bond with Thr233, which are considered to be significant for activity. The 3D representation of the human NLRP3 model constructed with 2D interactions of CY-09 at the binding site is shown in Figure 5. In addition, Tranilast interactions indicate that Thr233 and Tyr232 residues are required for the CY-09-like inhibitory activity. In addition, MCC950, a diarylsulfonylurea compound, has the second desired docking score in terms of binding affinity. Unlike CY-09, MCC950 has some interactions with various amino acid residues such as Arg314, Asp362, Glu363, and Arg406, mainly via Van der Waals and hydrogen bonds. On the other hand, the ligands Oridonin, MNS and OLT1177 have fewer interactions with related residues at the binding site and weaker binding affinities compared with CY-09, MCC950, and Tranilast.
The activation pathway of the NLRP3 inflammasome is an important process initiating cell pyroptosis in apical periodontitis or pulpitis [23]. When NLRP3 is activated, it associates with apoptosis-associated speck-like protein (ASC) and pro-caspase-1 to form the NLRP3-inflammatory complex [24]. ASC is a cytosolic protein composed of a C-terminal caspase recruitment domain (CARD) and a pyrin domain (PYD). NLRP3 interacts with ASC in the resulting complex. As a result, pro-caspase-1 and caspase-1 are activated, respectively. [24]. Caspase-1 activation cleaves pro-IL-1β and pro-IL-18 into their mature forms, IL-1β and IL-18 [24]. The active form of caspase-1 also causes the cleavage of Gasdermin D (GSDMD) at the N terminus [25]. Thus, GSDMD is an inflammatory caspase substrate responsible for the formation of pores in the cell membrane for pyroptotic cell death that occurs in apical periodontitis and pulpitis [25].
Pyroptosis, an inflammatory cell death, can occur due to the IL-1β release, and the effect of IL-1β production on pyroptotic processes was examined in a periodontal disease study [26]. As indicated in the NLRP3 inflammasome activation pathway (Figure 6), the accumulated IL-1β plays a key role in a number of inflammatory responses affecting the pathology of apical periodontitis and pulpitis. IL-1β can stimulate osteoclastogenesis through the kappa-B core ligand (RANKL) and expression of matrix metalloproteinases (MMPs) [27]. In addition to these effects, IL-1β stimulates the release of the pro-inflammatory mediators IL-6 and IL-8 from periodontal fibroblasts, leading to bone resorption [28]. Currently, in addition to the prevailing therapeutic strategies for apical periodontitis and pulpitis, the importance of NLRP3 inflammasome activation and caspase-1 inhibitors in endodontic treatments to treat IL-1β-related pyroptotic problems should be emphasized. NLRP3 inflammasome inhibitors, which contribute to IL-1β maturation, are currently used as a treatment for many inflammatory diseases [29]. However, studies on the effect of NLRP3 inhibitors on the apical periodontitis and pulpitis are rare. Previous studies have examined whether MCC950 blocks IL-1β and caspase-1 activation in mouse and human macrophages and have been shown to inhibit ASC oligomerization [29]. Peng et al. recently demonstrated that MCC950 downregulates the NLRP3 inflammasome signaling pathway in human periodontal ligament cells, thereby reducing the production of the pro-inflammatory cytokines IL-1β and IL-18, resulting in inhibition of osteogenesis [30]. Due to its specificity for NLRP3, MCC950 could be preferred for chronic and acute inflammation. As it reduces the production of IL-1β and IL-18, MCC950 can be considered as a potential therapeutic in the treatment of apical periodontitis and pulpitis. Furthermore, some selective caspase-1 inhibitors such as Belnacasan (VX-765) are currently used to block caspase-1 activity in inflammatory diseases, including periodontitis [31].
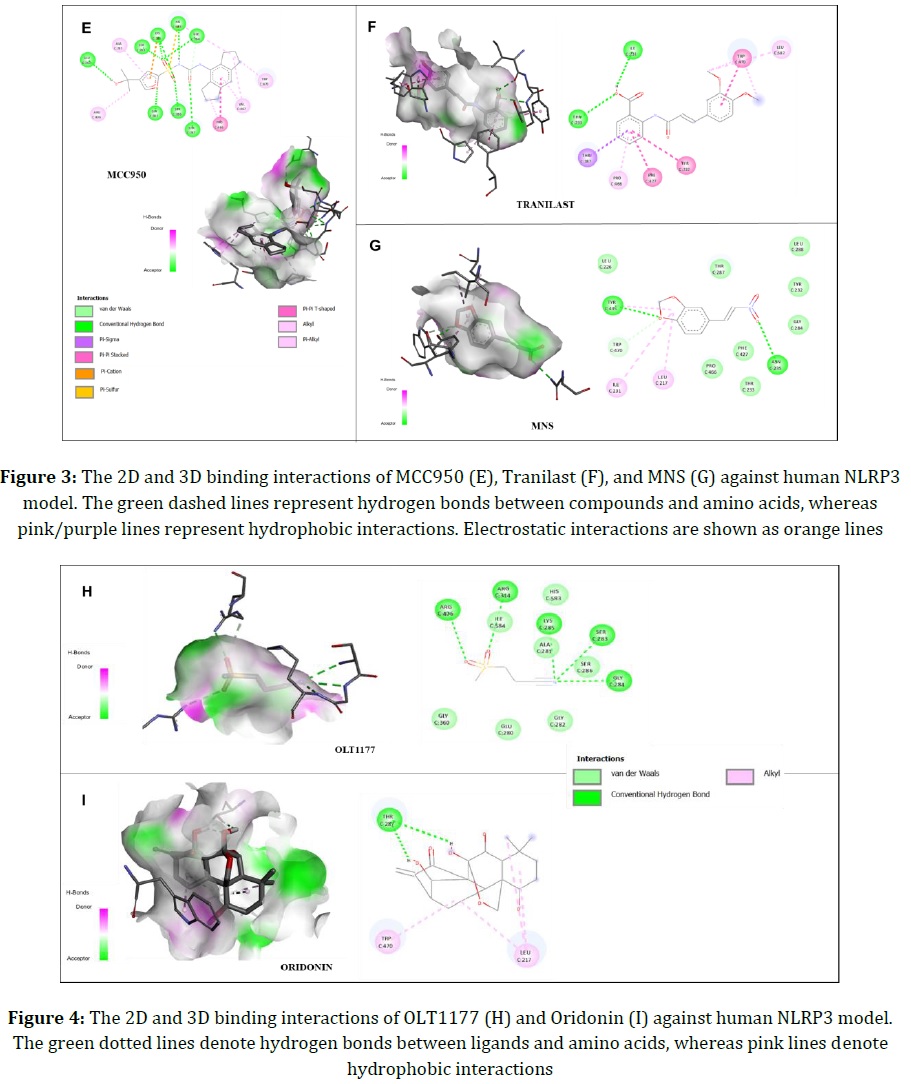
Concerning the previous studies on the NLRP3 inflammasome pathway, some NLRP3 and caspase-1 inhibitors were selected and a molecular docking study was performed for each ligand. The selected inhibitors were ranked and scored according to their target proteins. The interactions of the ligands with their target proteins, the binding free energies, and the inhibition constants were compared. The value of the inhibitor constant (Ki) is an indicator of determining the strength of a strong an inhibitor. Consistent with our results, ligands with higher binding affinities have lower inhibitor constant values. Therefore, low Ki values indicate that less drug is required for the inhibitory activity.
In a recent study, Moasses et al. examined the interfaces specified for the homo-interaction of the NLRP3-PYD domain [32]. Concerning previous in silico studies reported in the literature, the interacting amino acid residues found at each site were given as Ser5, Arg7, Cys8, Ala11, Glu15, Asp50, Val52, Asp53, and Thr56 [32]. Our results indicate that our ligands 16673-34-0 and BAY-117082 interact with active-site residues Ser5, Arg7, Cys8, and Thr56 of the PYD domain. These specified amino acid residues have been included in key residues for the type I site of the NLRP3-PYD domain. Therefore, they are expected to play an important role in the inhibitory activity of the selected ligands.
In the established human model of NLRP3, amino acid residues in the ADP-binding site overlap with residues previously reported to be important for the inhibitory activity [33]. These amino acid residues located at the ADP-binding site of the NOD2-ADP-bound structure (PDB: 5IRM) are Ile231, Tyr232, Thr233, Asn235, Ser283, Gly284, Lys285, Ser286, Thr287, Phe427, Pro466, and His58333. To inhibit the ATPase activity, Tyr232 and Thr233 residues present in the Walker A motif of the NACHT domain have been shown to be significant [33]. The CY-09 interactions with Tyr232 and Thr233 residues can be related to its high inhibitory activity. Tranilast, on the other hand, is a tryptophan analogue that binds directly to NLRP3 with similar binding interactions as CY-09. As demonstrated in a previous study, Asp362 and Glu363 are key residues in the Walker B motif of the NACHT domain, implying the inhibition of ATP hydrolysis [34]. Based on the results, the MCC950 interactions with Asp362 and Glu363 residues appear to be important to keep the NLRP3 inflammasome in an inactive state. Furthermore, Arg341 has been identified as a key amino acid residue for the structural domain of the caspase-1 protein-binding pocket [35]. The docking results of VX-765 and Parthenolide indicate that their interactions with residues Arg341, His342, and Arg383 are crucial for their activity. Hydrogen bonding between interacting residues and ligands was found to be significant for the inhibitory activity at the Caspase-1 active site. More importantly, VX-765 and Parthenolide formed bonds with almost the same amino acids in the binding pocket, indicating that they have similar binding and interaction modes. Therefore, we can conclude that the two drug molecules have similar inhibitory effects on caspase-1. However, VX-765 showed better binding affinity and lower Ki values than Partheolide, making VX-765 the preferred choice for use in inflammatory diseases of the endodontic diseases.
Conclusion
To sum up, the ligands CY-09 and VX-765 with the most desirable values in terms of binding affinity and inhibition constant can be used together in combination to prevent inflammation in the treatment of endodontic diseases. These inhibitors may help improve the viability of existing root canal medications and sealants, as well as pulp capping materials for future treatment of the endodontic infections. Furthermore, this study may shed light on the design and synthesis of novel NLRP3 inflammasome and caspase-1 inhibitors for use in LPS- and/or LTA-induced endodontic diseases.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Authors' contributions
All authors contributed to data analysis, drafting, and revising of the paper and agreed to be responsible for all the aspects of this work.
Conflict of Interest
We have no conflicts of interest to disclose.
Orcid:
Emine Erdag
https://orcid.org/0000-0002-1431-935X
HOW TO CITE THIS ARTICLE
Emine Erdag, Meltem Kucuk, Umut Aksoy, Nurettin Abacioglu, Ahmet Ozer Sehirli. Docking Study of Ligands Targeting NLRP3 Inflammatory Pathway for Endodontic Diseases. Chem. Methodol., 2023, 7(3) 200-210


